| Namespace | http://www.openmicroscopy.org/Schemas/SPW/2013-06 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Annotations
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
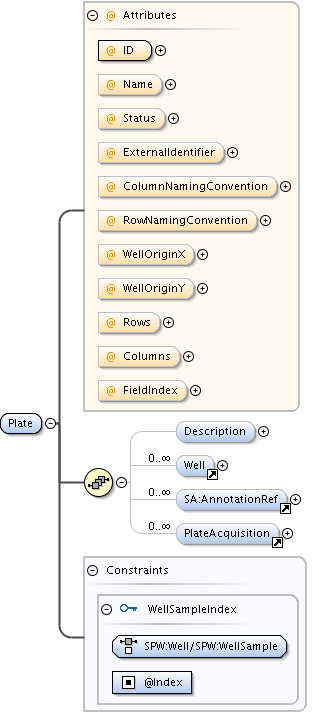
Diagram
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Properties
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Used by
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
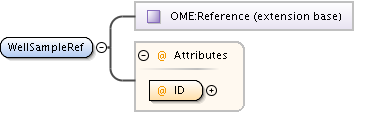
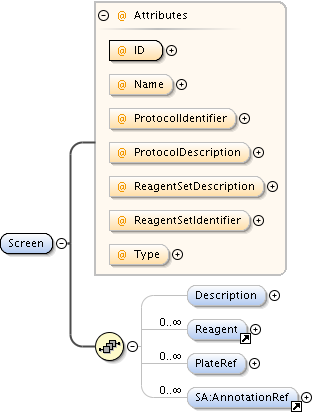
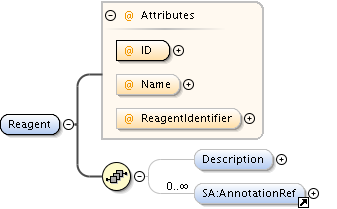
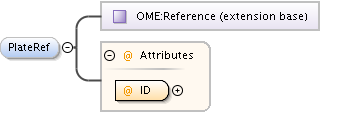
| Model | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
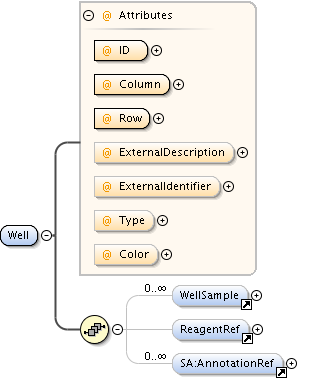
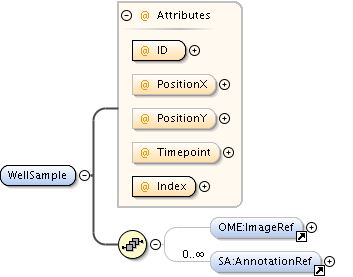
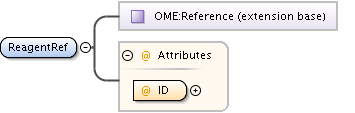
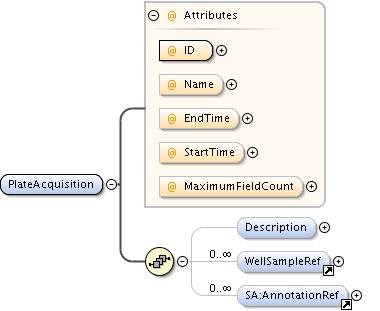
| Children | AnnotationRef, Description, PlateAcquisition, Well | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Instance
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Attributes
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Source
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Schema location | http://www.openmicroscopy.org/Schemas/SPW/2013-06/SPW.xsd |