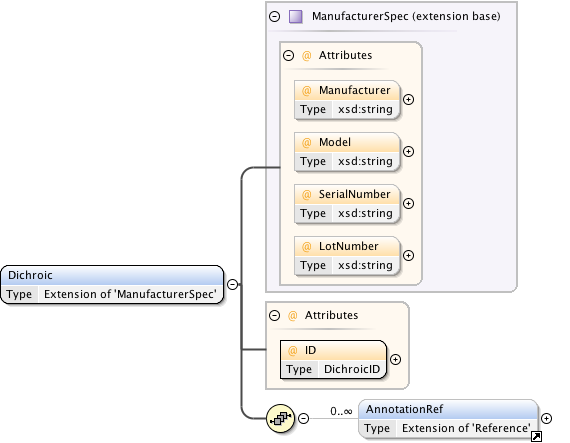
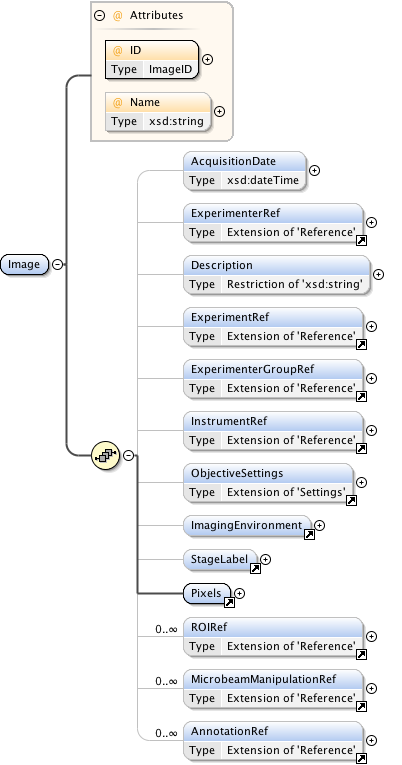
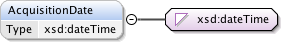
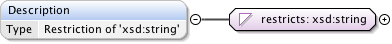
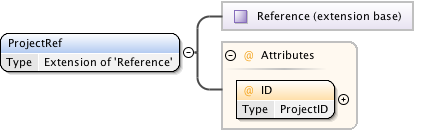
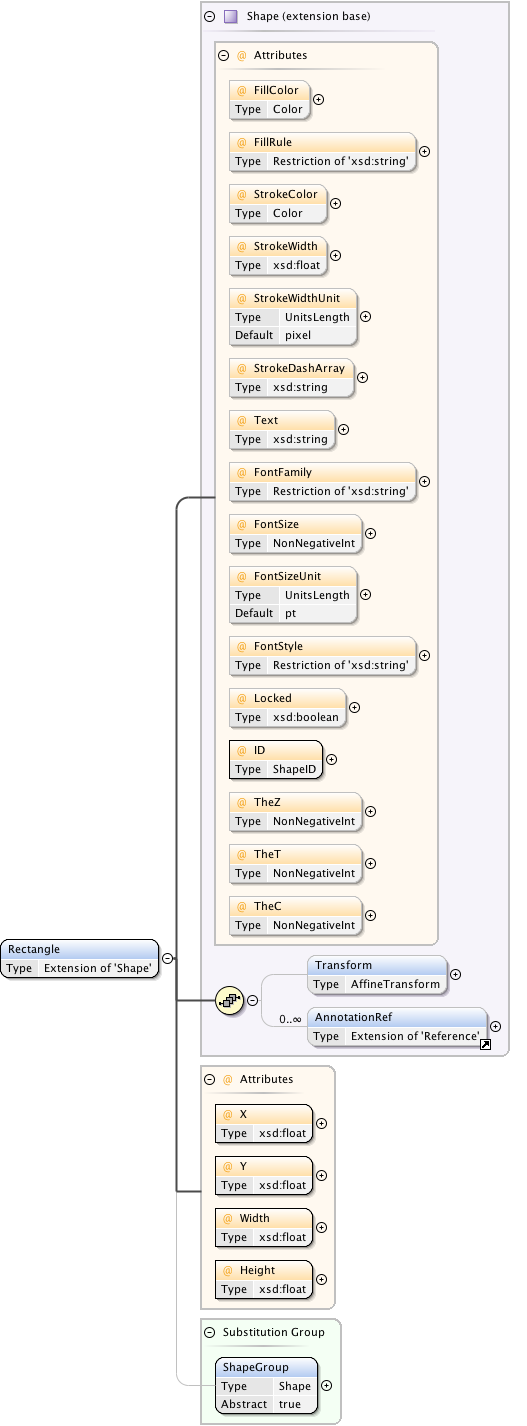
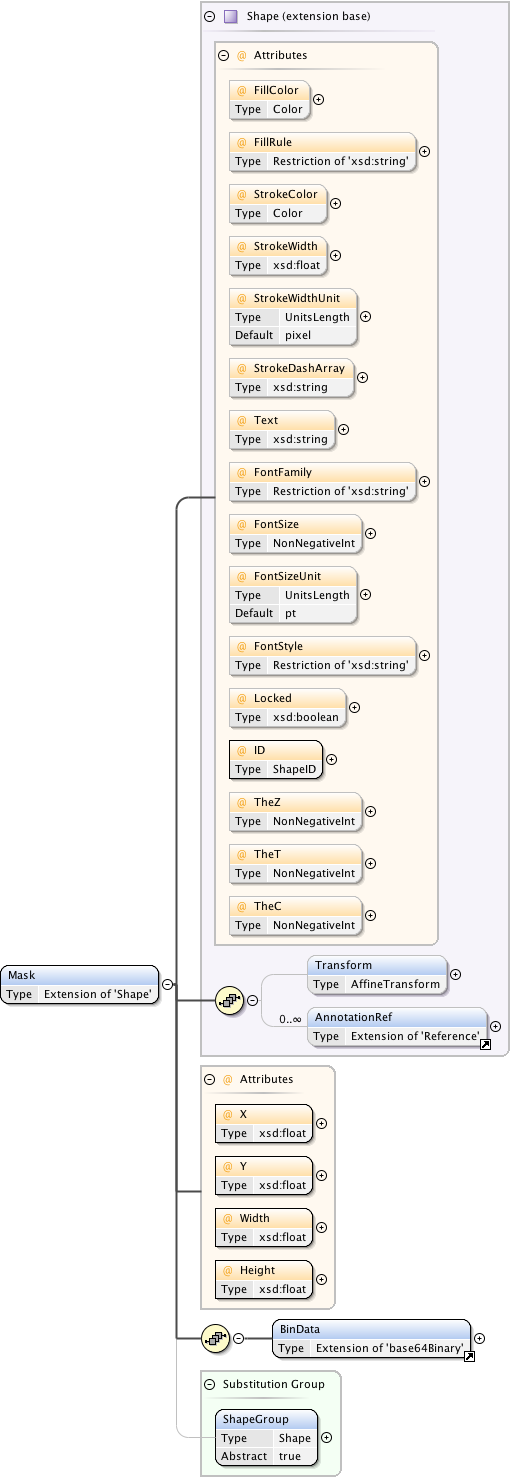
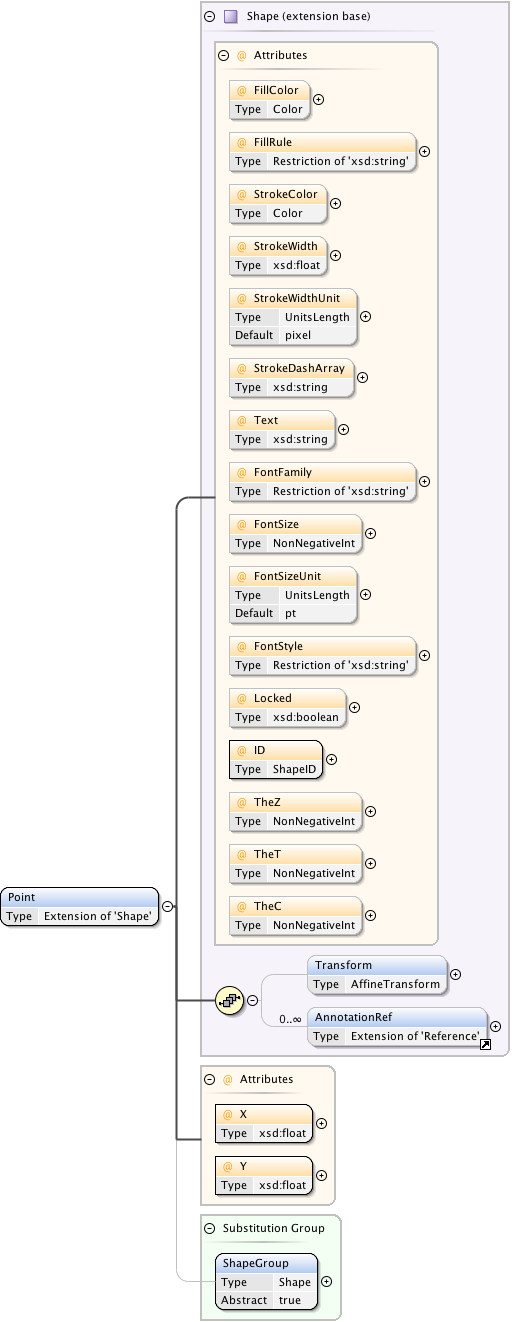
| Namespace | http://www.openmicroscopy.org/Schemas/OME/2016-06 | ||||||||||||||
|
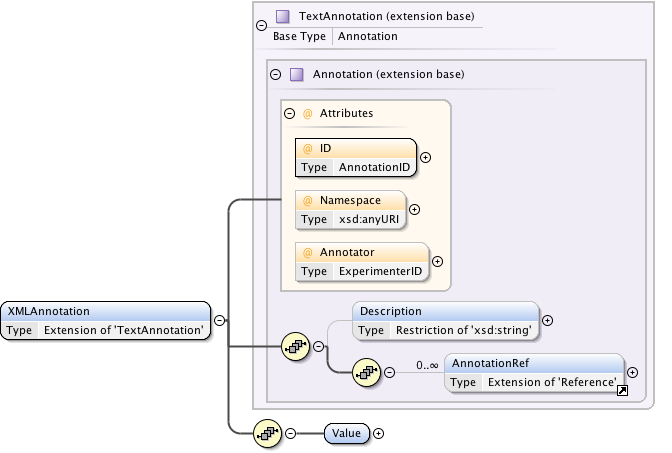
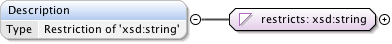
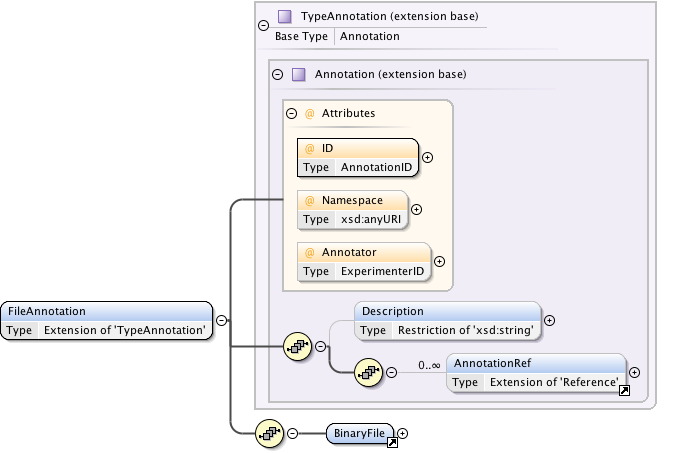
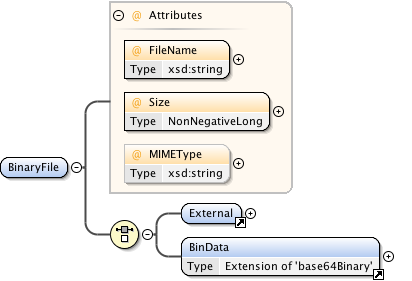
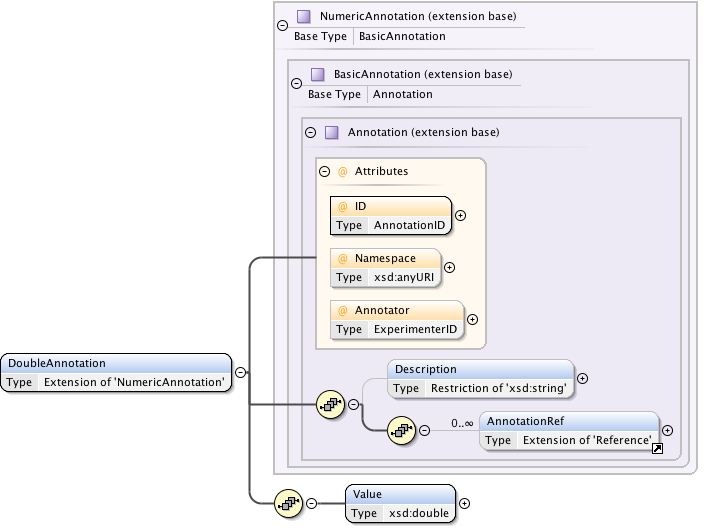
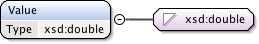
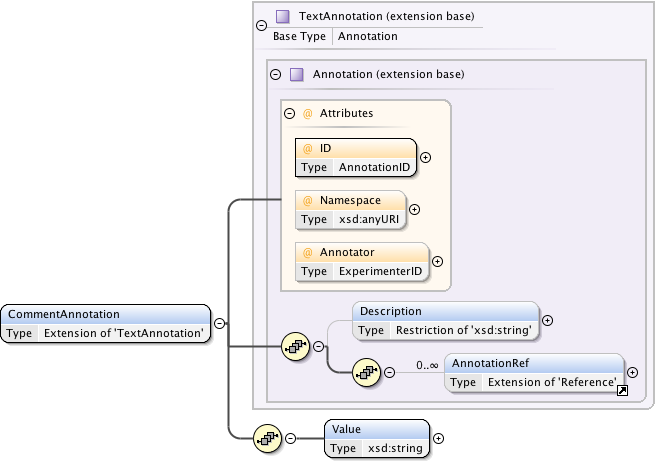
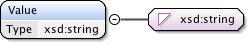
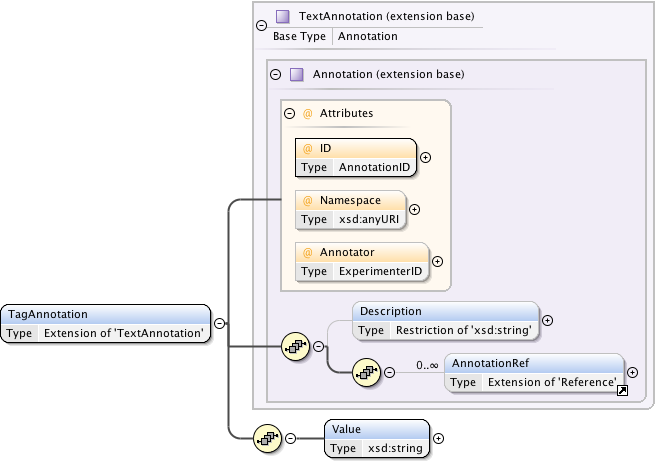
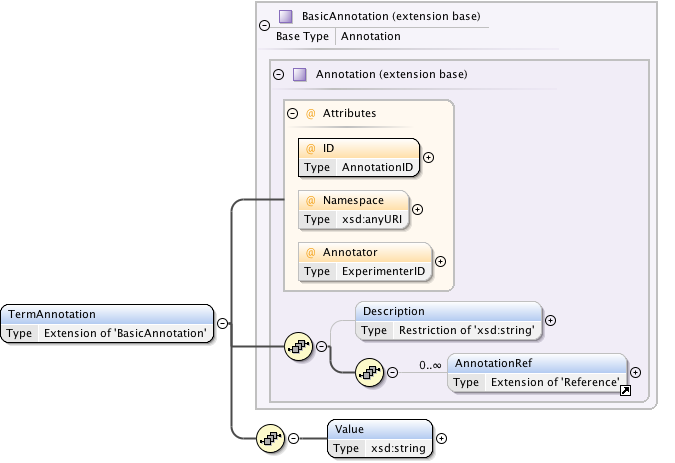
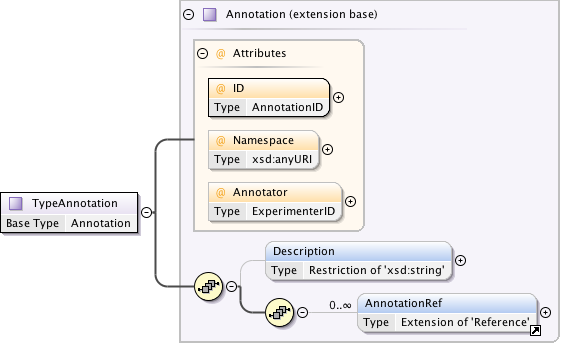
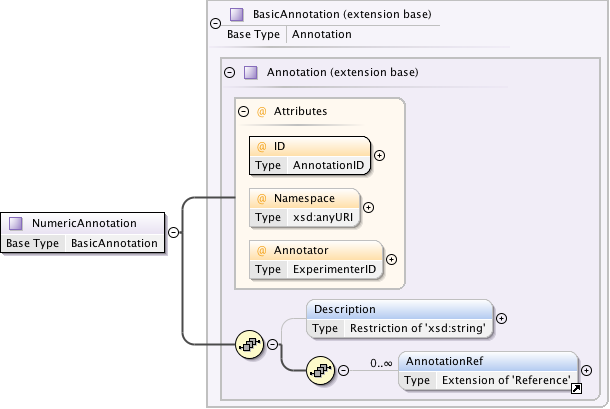
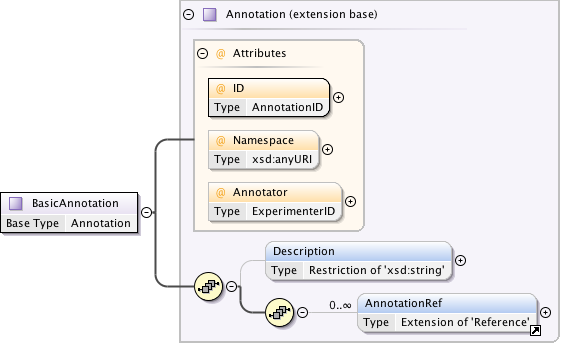
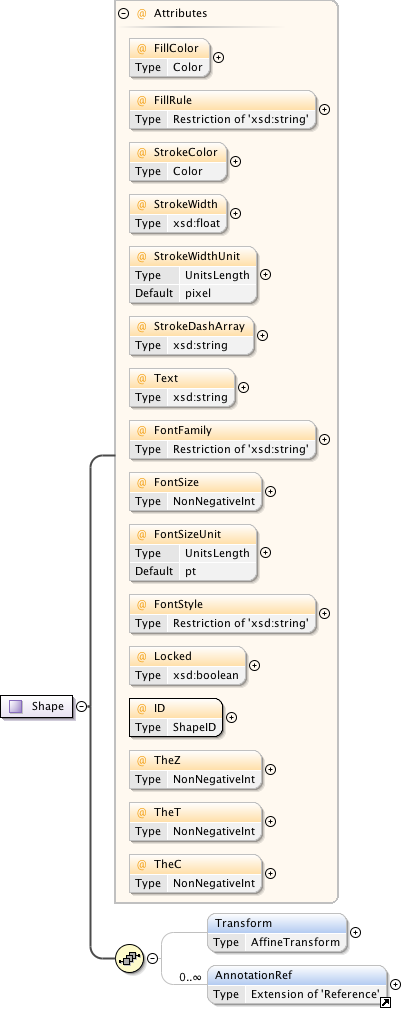
Annotations
|
|
||||||||||||||
|
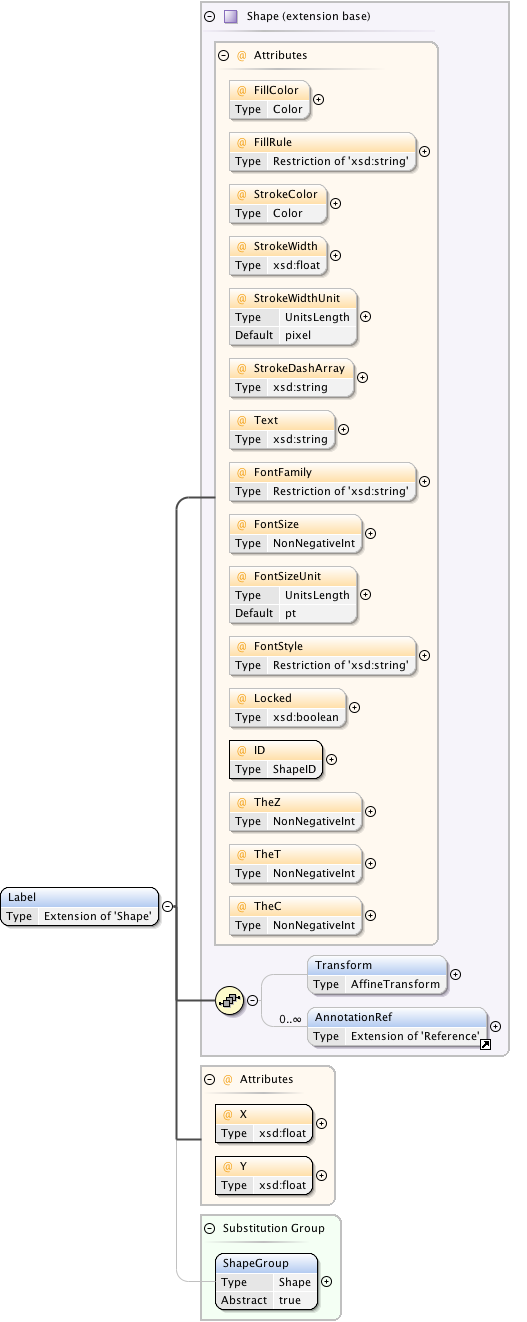
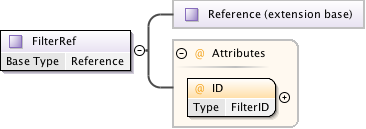
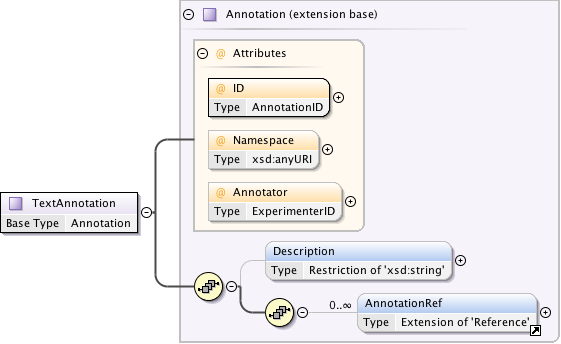
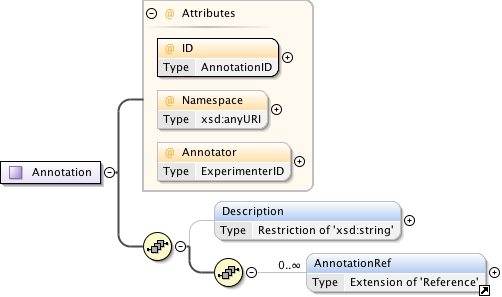
Diagram
|
 |
||||||||||||||
|
Properties
|
|
||||||||||||||
| Model |
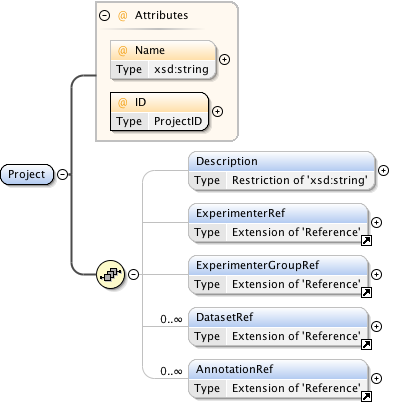
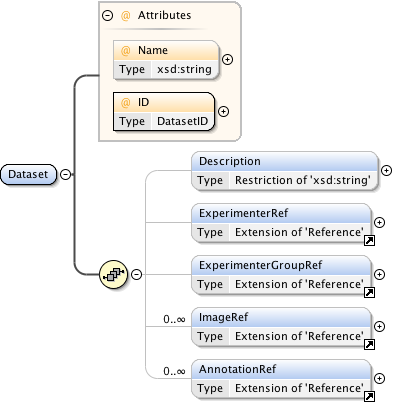
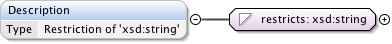
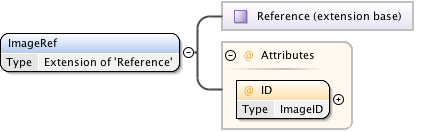
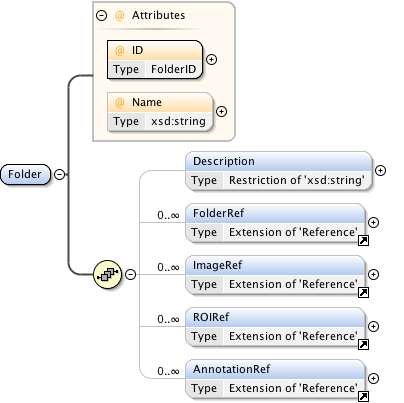
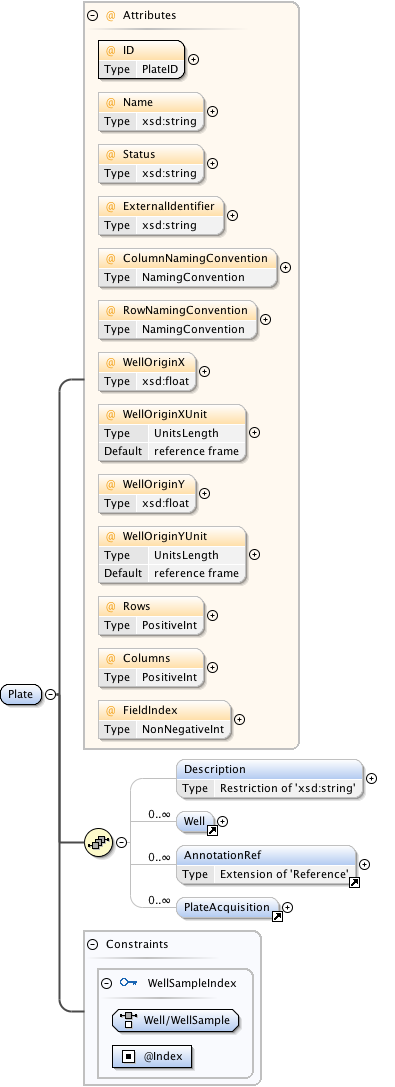
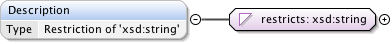
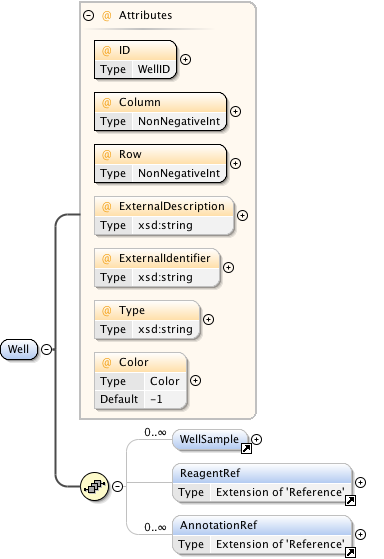
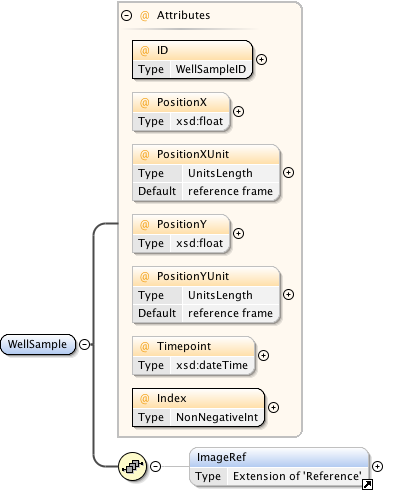
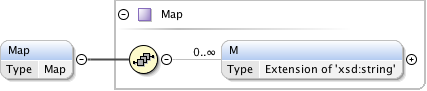
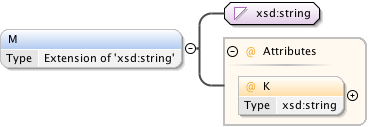
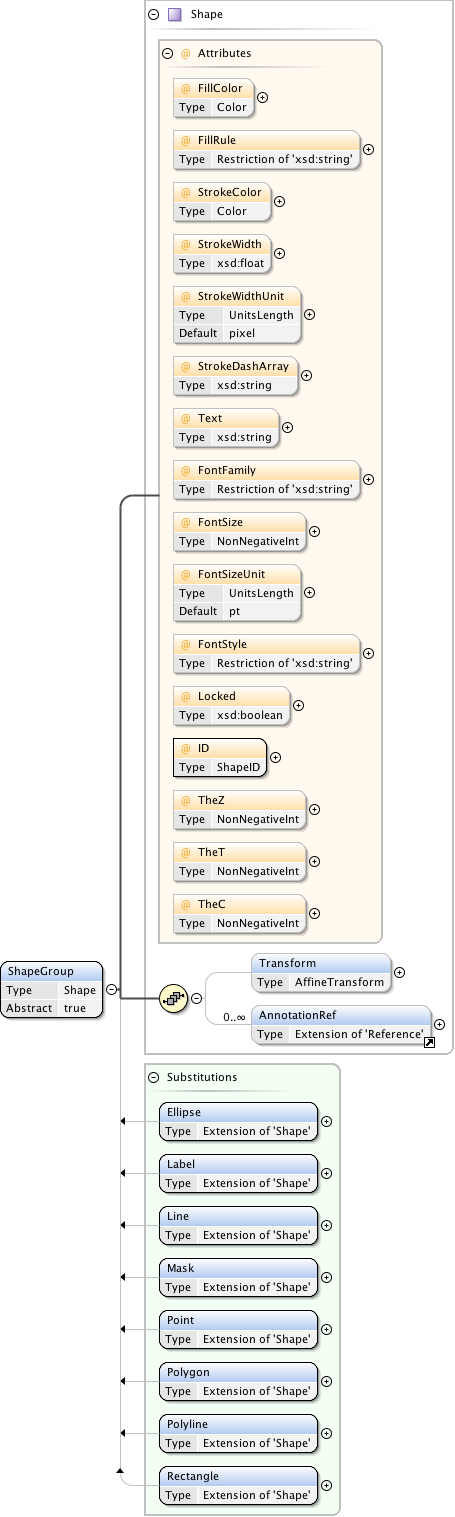
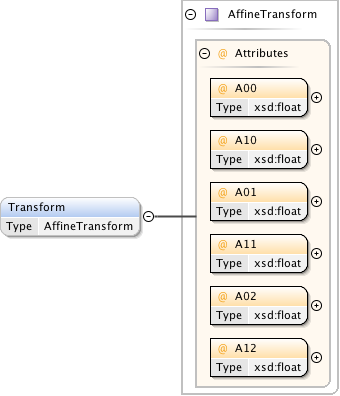
Rights{0,1} , ((Project* , Dataset* , Folder* , Experiment* , Plate* , Screen* , Experimenter* , ExperimenterGroup* , Instrument* , Image* , StructuredAnnotations{0,1} , ROI*) | BinaryOnly)
|
||||||||||||||
| Children | BinaryOnly, Dataset, Experiment, Experimenter, ExperimenterGroup, Folder, Image, Instrument, Plate, Project, ROI, Rights, Screen, StructuredAnnotations | ||||||||||||||
|
Instance
|
|
||||||||||||||
|
Attributes
|
|
||||||||||||||
|
Source
|
|
||||||||||||||
| Schema location | http://www.openmicroscopy.org/Schemas/OME/2016-06/ome.xsd |