Designing an open multi-resolution file format
In 2012, the OME Consortium started working on the support of multi-resolution, or pyramidal, proprietary file formats. The result of this work was the addition of several proprietary whole slide image (WSI) file format readers in Bio-Formats releases during 2013-2017. As often happens, proprietary file format (PFF) readers are only part of the solution required by the scientific and clinical communities - see also our blog post. We have been repeatedly asked to build an open, supported alternative for storing large WSIs. For some time, we have deferred to the work in DICOM Supplement 145 introduced in 2010, but this work has not produced viable, supported, open implementations nor considered the rapidly emerging field of multiplexed WSIs. Therefore, we started working on an extension to the OME-TIFF format specification to support pyramidal resolutions at the beginning of 2018. This scoping exercise resulted in a formal open proposal announced to the community in February 2018.
Extending a widely adopted format specification with hundreds of instruments generating files in production is not without risk. Compatibility between the various formats and software version is usually a key target of the process. The principal challenges are:
- maintaining the usability of new files using either older versions of OME software or third-party software
- minimizing the risks of conflicts of the proposed extension with other specifications
The OME team has reviewed several approaches for representing multi-resolution pyramidal levels in TIFF files. In the end, the solution we chose offered the maximum compatibility with existing reference tools while remaining unambiguous and compliant with the official TIFF specification.
Reading and writing OME-TIFF pyramids
Following the decisions on the formats specification, we began working to enable support for reading and writing OME-TIFF pyramids in our Java-based application stack, specifically, Bio-Formats and OMERO.
We demonstrated the first part of this work during the 2018 OME Users Meeting in Dundee. We presented example OME-TIFF files derived from public pyramidal files acquired using the Leica software. Furthermore, these files were imported into an OMERO server.
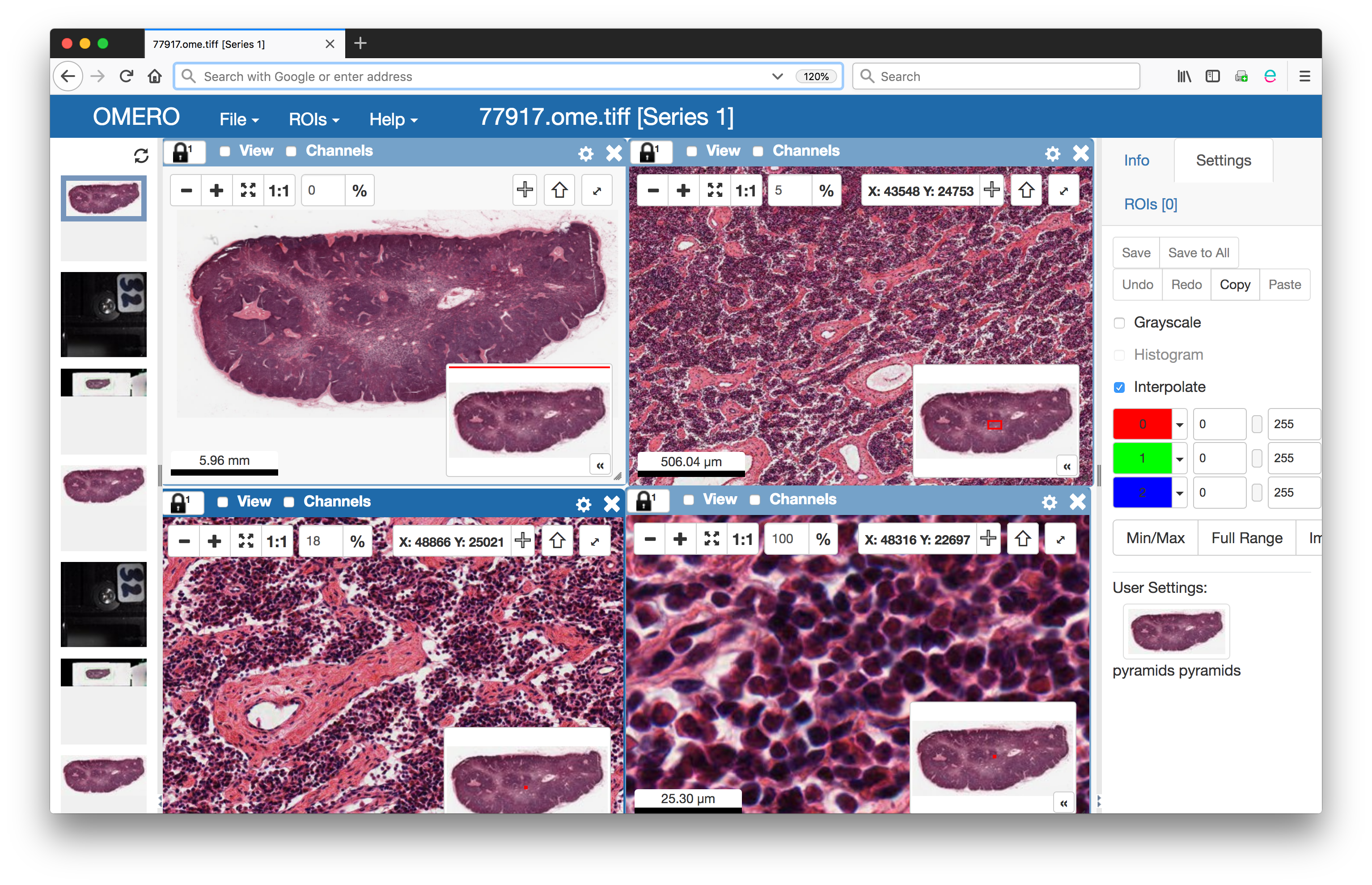 WSIs stored as pyramidal OME-TIFF explored visually using
OMERO.iviewer Web viewer
WSIs stored as pyramidal OME-TIFF explored visually using
OMERO.iviewer Web viewer
Since then, we focused our efforts on the generation of pyramidal OME-TIFF files using Bio-Formats. As of today, we are proud to have a milestone release of Bio-Formats available here with full support for reading and writing open OME-TIFF files with support for pyramids. The software allows both the conversion of existing proprietary file formats as well as the generation of pyramidal levels from large resolution planes.
Towards Bio-Formats 6.0.0 and beyond
What are our next steps? We first require more testing to ensure that the changes have not affected our existing image reading library and are compatible with Bio-Formats’ existing support for pyramidal file formats. This includes several tens of file formats generated by manufacturers like Leica/Aperio, Hamamatsu, and Zeiss.
We are also working on ongoing performance improvements to ensure consumers can write compliant OME-TIFF rapidly either using the API directly or via our end-users tools like the Bio-Formats command-line utility, the ImageJ plugin or the OMERO exporter.
Finally, we will review the API changes necessary so that Bio-Formats can read and write compliant OME-TIFF pyramids. We have maintained our API fully backwards-compatible for the last two years following our last release of the OME model. At present, we believe a new major version of Bio-Formats 6.0.0 including breaking API changes will be released by the end of 2018. An OMERO release that supports Bio-Formats 6.0.0 will follow, but the exact date is not yet known. For those of you following our develop branch, you will see IDR consuming Bio-Formats 6.0.0 not too long after release.
Once this work is completed, our next aims in terms of formats will be:
- Continue the work on OME formats using new binary vessels also presented at the 2018 Users Meeting. Bio-Formats 6.0.0-m3 already includes a reader for the Keller Lab Block format and Bio-Formats 6 will include a reader for the Big DataViewer format.
- Work with the community to keep OME-Files C++ support for pyramidal OME-TIFF in sync with the Java changes - read this ome-users thread to know more about recent improvements driven by the community.

